Cell biology
| Part of a series on |
| Biology |
|---|
Cell biology (also cellular biology or cytology) is a branch of biology that studies the structure, function, and behavior of cells.[1][2] All living organisms are made of cells. A cell is the basic unit of life that is responsible for the living and functioning of organisms.[3] Cell biology is the study of the structural and functional units of cells. Cell biology encompasses both prokaryotic and eukaryotic cells and has many subtopics which may include the study of cell metabolism, cell communication, cell cycle, biochemistry, and cell composition. The study of cells is performed using several microscopy techniques, cell culture, and cell fractionation. These have allowed for and are currently being used for discoveries and research pertaining to how cells function, ultimately giving insight into understanding larger organisms. Knowing the components of cells and how cells work is fundamental to all biological sciences while also being essential for research in biomedical fields such as cancer, and other diseases. Research in cell biology is interconnected to other fields such as genetics, molecular genetics, molecular biology, medical microbiology, immunology, and cytochemistry.
History
[edit]Cells were first seen in 17th-century Europe with the invention of the compound microscope. In 1665, Robert Hooke referred to the building blocks of all living organisms as "cells" (published in Micrographia) after looking at a piece of cork and observing a structure reminiscent of a monastic cell;[4][5] however, the cells were dead. They gave no indication to the actual overall components of a cell. A few years later, in 1674, Anton Van Leeuwenhoek was the first to analyze live cells in his examination of algae. Many years later, in 1831, Robert Brown discovered the nucleus. All of this preceded the cell theory which states that all living things are made up of cells and that cells are organisms' functional and structural units. This was ultimately concluded by plant scientist Matthias Schleiden[5] and animal scientist Theodor Schwann in 1838, who viewed live cells in plant and animal tissue, respectively.[3] 19 years later, Rudolf Virchow further contributed to the cell theory, adding that all cells come from the division of pre-existing cells.[3] Viruses are not considered in cell biology – they lack the characteristics of a living cell and instead are studied in the microbiology subclass of virology.[6]
Techniques
[edit]Cell biology research looks at different ways to culture and manipulate cells outside of a living body to further research in human anatomy and physiology, and to derive medications. The techniques by which cells are studied have evolved. Due to advancements in microscopy, techniques and technology have allowed scientists to hold a better understanding of the structure and function of cells. Many techniques commonly used to study cell biology are listed below:[7]
- Cell culture: Utilizes rapidly growing cells on media which allows for a large amount of a specific cell type and an efficient way to study cells.[8] Cell culture is one of the major tools used in cellular and molecular biology, providing excellent model systems for studying the normal physiology and biochemistry of cells (e.g., metabolic studies, aging), the effects of drugs and toxic compounds on the cells, and mutagenesis and carcinogenesis. It is also used in drug screening and development, and large scale manufacturing of biological compounds (e.g., vaccines, therapeutic proteins).
- Fluorescence microscopy: Fluorescent markers such as GFP, are used to label a specific component of the cell. Afterwards, a certain light wavelength is used to excite the fluorescent marker which can then be visualized.[8]
- Phase-contrast microscopy: Uses the optical aspect of light to represent the solid, liquid, and gas-phase changes as brightness differences.[8]
- Confocal microscopy: Combines fluorescence microscopy with imaging by focusing light and snap shooting instances to form a 3-D image.[8]
- Transmission electron microscopy: Involves metal staining and the passing of electrons through the cells, which will be deflected upon interaction with metal. This ultimately forms an image of the components being studied.[8]
- Cytometry: The cells are placed in the machine which uses a beam to scatter the cells based on different aspects and can therefore separate them based on size and content. Cells may also be tagged with GFP-fluorescence and can be separated that way as well.[9]
- Cell fractionation: This process requires breaking up the cell using high temperature or sonification followed by centrifugation to separate the parts of the cell allowing for them to be studied separately.[8]
Cell types
[edit]
There are two fundamental classifications of cells: prokaryotic and eukaryotic. Prokaryotic cells are distinguished from eukaryotic cells by the absence of a cell nucleus or other membrane-bound organelle.[10] Prokaryotic cells are much smaller than eukaryotic cells, making them the smallest form of life.[11] Prokaryotic cells include Bacteria and Archaea, and lack an enclosed cell nucleus. Eukaryotic cells are found in plants, animals, fungi, and protists. They range from 10 to 100 μm in diameter, and their DNA is contained within a membrane-bound nucleus. Eukaryotes are organisms containing eukaryotic cells. The four eukaryotic kingdoms are Animalia, Plantae, Fungi, and Protista.[12]
They both reproduce through binary fission. Bacteria, the most prominent type, have several different shapes, although most are spherical or rod-shaped. Bacteria can be classed as either gram-positive or gram-negative depending on the cell wall composition. Gram-positive bacteria have a thicker peptidoglycan layer than gram-negative bacteria. Bacterial structural features include a flagellum that helps the cell to move,[13] ribosomes for the translation of RNA to protein,[13] and a nucleoid that holds all the genetic material in a circular structure.[13] There are many processes that occur in prokaryotic cells that allow them to survive. In prokaryotes, mRNA synthesis is initiated at a promoter sequence on the DNA template comprising two consensus sequences that recruit RNA polymerase. The prokaryotic polymerase consists of a core enzyme of four protein subunits and a σ protein that assists only with initiation. For instance, in a process termed conjugation, the fertility factor allows the bacteria to possess a pilus which allows it to transmit DNA to another bacteria which lacks the F factor, permitting the transmittance of resistance allowing it to survive in certain environments.[14]
Structure and function
[edit]Structure of eukaryotic cells
[edit]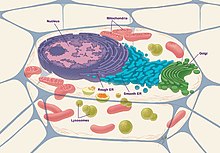
Eukaryotic cells are composed of the following organelles:
- Nucleus: The nucleus of the cell functions as the genome and genetic information storage for the cell, containing all the DNA organized in the form of chromosomes. It is surrounded by a nuclear envelope, which includes nuclear pores allowing for the transportation of proteins between the inside and outside of the nucleus.[15] This is also the site for replication of DNA as well as transcription of DNA to RNA. Afterwards, the RNA is modified and transported out to the cytosol to be translated to protein.[16]
- Nucleolus: This structure is within the nucleus, usually dense and spherical. It is the site of ribosomal RNA (rRNA) synthesis, which is needed for ribosomal assembly.
- Endoplasmic reticulum (ER): This functions to synthesize, store, and secrete proteins to the Golgi apparatus.[17] Structurally, the endoplasmic reticulum is a network of membranes found throughout the cell and connected to the nucleus. The membranes are slightly different from cell to cell and a cell's function determines the size and structure of the ER.[18]
- Mitochondria: Commonly known as the powerhouse of the cell is a double membrane bound cell organelle.[19] This functions for the production of energy or ATP within the cell. Specifically, this is the place where the Krebs cycle or TCA cycle for the production of NADH and FADH occurs. Afterwards, these products are used within the electron transport chain (ETC) and oxidative phosphorylation for the final production of ATP.[20]
- Golgi apparatus: This functions to further process, package, and secrete the proteins to their destination. The proteins contain a signal sequence that allows the Golgi apparatus to recognize and direct it to the correct place. Golgi apparatus also produce glycoproteins and glycolipids.[21]
- Lysosome: The lysosome functions to degrade material brought in from the outside of the cell or old organelles. This contains many acid hydrolases, proteases, nucleases, and lipases, which break down the various molecules. Autophagy is the process of degradation through lysosomes which occurs when a vesicle buds off from the ER and engulfs the material, then, attaches and fuses with the lysosome to allow the material to be degraded.[22]
- Ribosomes: Functions to translate RNA to protein. it serves as a site of protein synthesis.[23]
- Cytoskeleton: Cytoskeleton is a structure that helps to maintain the shape and general organization of the cytoplasm. It anchors organelles within the cells and makes up the structure and stability of the cell. The cytoskeleton is composed of three principal types of protein filaments: actin filaments, intermediate filaments, and microtubules, which are held together and linked to subcellular organelles and the plasma membrane by a variety of accessory proteins.[24]
- Cell membrane: The cell membrane can be described as a phospholipid bilayer and is also consisted of lipids and proteins.[13] Because the inside of the bilayer is hydrophobic and in order for molecules to participate in reactions within the cell, they need to be able to cross this membrane layer to get into the cell via osmotic pressure, diffusion, concentration gradients, and membrane channels.[25]
- Centrioles: Function to produce spindle fibers which are used to separate chromosomes during cell division.
Eukaryotic cells may also be composed of the following molecular components:
- Chromatin: This makes up chromosomes and is a mixture of DNA with various proteins.
- Cilia: They help to propel substances and can also be used for sensory purposes.[26]
Cell metabolism
[edit]Cell metabolism is necessary for the production of energy for the cell and therefore its survival and includes many pathways and also sustaining the main cell organelles such as the nucleus, the mitochondria, the cell membrane etc. For cellular respiration, once glucose is available, glycolysis occurs within the cytosol of the cell to produce pyruvate. Pyruvate undergoes decarboxylation using the multi-enzyme complex to form acetyl coA which can readily be used in the TCA cycle to produce NADH and FADH2. These products are involved in the electron transport chain to ultimately form a proton gradient across the inner mitochondrial membrane. This gradient can then drive the production of ATP and H2O during oxidative phosphorylation.[27] Metabolism in plant cells includes photosynthesis which is simply the exact opposite of respiration as it ultimately produces molecules of glucose.
Cell signaling
[edit]Cell signaling or cell communication is important for cell regulation and for cells to process information from the environment and respond accordingly. Signaling can occur through direct cell contact or endocrine, paracrine, and autocrine signaling. Direct cell-cell contact is when a receptor on a cell binds a molecule that is attached to the membrane of another cell. Endocrine signaling occurs through molecules secreted into the bloodstream. Paracrine signaling uses molecules diffusing between two cells to communicate. Autocrine is a cell sending a signal to itself by secreting a molecule that binds to a receptor on its surface. Forms of communication can be through:
- Ion channels: Can be of different types such as voltage or ligand gated ion channels. They allow for the outflow and inflow of molecules and ions.
- G-protein coupled receptor (GPCR): Is widely recognized to contain seven transmembrane domains. The ligand binds on the extracellular domain and once the ligand binds, this signals a guanine exchange factor to convert GDP to GTP and activate the G-α subunit. G-α can target other proteins such as adenyl cyclase or phospholipase C, which ultimately produce secondary messengers such as cAMP, Ip3, DAG, and calcium. These secondary messengers function to amplify signals and can target ion channels or other enzymes. One example for amplification of a signal is cAMP binding to and activating PKA by removing the regulatory subunits and releasing the catalytic subunit. The catalytic subunit has a nuclear localization sequence which prompts it to go into the nucleus and phosphorylate other proteins to either repress or activate gene activity.[27]
- Receptor tyrosine kinases: Bind growth factors, further promoting the tyrosine on the intracellular portion of the protein to cross phosphorylate. The phosphorylated tyrosine becomes a landing pad for proteins containing an SH2 domain allowing for the activation of Ras and the involvement of the MAP kinase pathway.[28]
Growth and development
[edit]Eukaryotic cell cycle
[edit]
Cells are the foundation of all organisms and are the fundamental units of life. The growth and development of cells are essential for the maintenance of the host and survival of the organism. For this process, the cell goes through the steps of the cell cycle and development which involves cell growth, DNA replication, cell division, regeneration, and cell death.
The cell cycle is divided into four distinct phases: G1, S, G2, and M. The G phase – which is the cell growth phase – makes up approximately 95% of the cycle. The proliferation of cells is instigated by progenitors. All cells start out in an identical form and can essentially become any type of cells. Cell signaling such as induction can influence nearby cells to determinate the type of cell it will become. Moreover, this allows cells of the same type to aggregate and form tissues, then organs, and ultimately systems. The G1, G2, and S phase (DNA replication, damage and repair) are considered to be the interphase portion of the cycle, while the M phase (mitosis) is the cell division portion of the cycle. Mitosis is composed of many stages which include, prophase, metaphase, anaphase, telophase, and cytokinesis, respectively. The ultimate result of mitosis is the formation of two identical daughter cells.
The cell cycle is regulated in cell cycle checkpoints, by a series of signaling factors and complexes such as cyclins, cyclin-dependent kinase, and p53. When the cell has completed its growth process and if it is found to be damaged or altered, it undergoes cell death, either by apoptosis or necrosis, to eliminate the threat it can cause to the organism's survival.[29]
Cell mortality, cell lineage immortality
[edit]The ancestry of each present day cell presumably traces back, in an unbroken lineage for over 3 billion years to the origin of life. It is not actually cells that are immortal but multi-generational cell lineages.[30] The immortality of a cell lineage depends on the maintenance of cell division potential. This potential may be lost in any particular lineage because of cell damage, terminal differentiation as occurs in nerve cells, or programmed cell death (apoptosis) during development. Maintenance of cell division potential over successive generations depends on the avoidance and the accurate repair of cellular damage, particularly DNA damage. In sexual organisms, continuity of the germline depends on the effectiveness of processes for avoiding DNA damage and repairing those DNA damages that do occur. Sexual processes in eukaryotes, as well as in prokaryotes, provide an opportunity for effective repair of DNA damages in the germ line by homologous recombination.[30][31]
Cell cycle phases
[edit]The cell cycle is a four-stage process that a cell goes through as it develops and divides. It includes Gap 1 (G1), synthesis (S), Gap 2 (G2), and mitosis (M). The cell either restarts the cycle from G1 or leaves the cycle through G0 after completing the cycle. The cell can progress from G0 through terminal differentiation. Finally, the interphase refers to the phases of the cell cycle that occur between one mitosis and the next, and includes G1, S, and G2. Thus, the phases are:
- G1 phase: the cell grows in size and its contents are replicated.
- S phase: the cell replicates each of the 46 chromosomes.
- G2 phase: in preparation for cell division, new organelles and proteins form.
- M phase: cytokinesis occurs, resulting in two identical daughter cells.
- G0 phase: the two cells enter a resting stage where they do their job without actively preparing to divide.[32]
Pathology
[edit]The scientific branch that studies and diagnoses diseases on the cellular level is called cytopathology. Cytopathology is generally used on samples of free cells or tissue fragments, in contrast to the pathology branch of histopathology, which studies whole tissues. Cytopathology is commonly used to investigate diseases involving a wide range of body sites, often to aid in the diagnosis of cancer but also in the diagnosis of some infectious diseases and other inflammatory conditions. For example, a common application of cytopathology is the Pap smear, a screening test used to detect cervical cancer, and precancerous cervical lesions that may lead to cervical cancer.[33]
Cell cycle checkpoints and DNA damage repair system
[edit]The cell cycle is composed of a number of well-ordered, consecutive stages that result in cellular division. The fact that cells do not begin the next stage until the last one is finished, is a significant element of cell cycle regulation. Cell cycle checkpoints are characteristics that constitute an excellent monitoring strategy for accurate cell cycle and divisions. Cdks, associated cyclin counterparts, protein kinases, and phosphatases regulate cell growth and division from one stage to another.[34] The cell cycle is controlled by the temporal activation of Cdks, which is governed by cyclin partner interaction, phosphorylation by particular protein kinases, and de-phosphorylation by Cdc25 family phosphatases. In response to DNA damage, a cell's DNA repair reaction is a cascade of signaling pathways that leads to checkpoint engagement, regulates, the repairing mechanism in DNA, cell cycle alterations, and apoptosis. Numerous biochemical structures, as well as processes that detect damage in DNA, are ATM and ATR, which induce the DNA repair checkpoints[35]
The cell cycle is a sequence of activities in which cell organelles are duplicated and subsequently separated into daughter cells with precision. There are major events that happen during a cell cycle. The processes that happen in the cell cycle include cell development, replication and segregation of chromosomes. The cell cycle checkpoints are surveillance systems that keep track of the cell cycle's integrity, accuracy, and chronology. Each checkpoint serves as an alternative cell cycle endpoint, wherein the cell's parameters are examined and only when desirable characteristics are fulfilled does the cell cycle advance through the distinct steps. The cell cycle's goal is to precisely copy each organism's DNA and afterwards equally split the cell and its components between the two new cells. Four main stages occur in the eukaryotes. In G1, the cell is usually active and continues to grow rapidly, while in G2, the cell growth continues while protein molecules become ready for separation. These are not dormant times; they are when cells gain mass, integrate growth factor receptors, establish a replicated genome, and prepare for chromosome segregation. DNA replication is restricted to a separate Synthesis in eukaryotes, which is also known as the S-phase. During mitosis, which is also known as the M-phase, the segregation of the chromosomes occur.[36] DNA, like every other molecule, is capable of undergoing a wide range of chemical reactions. Modifications in DNA's sequence, on the other hand, have a considerably bigger impact than modifications in other cellular constituents like RNAs or proteins because DNA acts as a permanent copy of the cell genome. When erroneous nucleotides are incorporated during DNA replication, mutations can occur. The majority of DNA damage is fixed by removing the defective bases and then re-synthesizing the excised area. On the other hand, some DNA lesions can be mended by reversing the damage, which may be a more effective method of coping with common types of DNA damage. Only a few forms of DNA damage are mended in this fashion, including pyrimidine dimers caused by ultraviolet (UV) light changed by the insertion of methyl or ethyl groups at the purine ring's O6 position.[37]
Mitochondrial membrane dynamics
[edit]Mitochondria are commonly referred to as the cell's "powerhouses" because of their capacity to effectively produce ATP which is essential to maintain cellular homeostasis and metabolism. Moreover, researchers have gained a better knowledge of mitochondria's significance in cell biology because of the discovery of cell signaling pathways by mitochondria which are crucial platforms for cell function regulation such as apoptosis. Its physiological adaptability is strongly linked to the cell mitochondrial channel's ongoing reconfiguration through a range of mechanisms known as mitochondrial membrane dynamics, including endomembrane fusion and fragmentation (separation) and ultrastructural membrane remodeling. As a result, mitochondrial dynamics regulate and frequently choreograph not only metabolic but also complicated cell signaling processes such as cell pluripotent stem cells, proliferation, maturation, aging, and mortality. Mutually, post-translational alterations of mitochondrial apparatus and the development of transmembrane contact sites among mitochondria and other structures, which both have the potential to link signals from diverse routes that affect mitochondrial membrane dynamics substantially,[36] Mitochondria are wrapped by two membranes: an inner mitochondrial membrane (IMM) and an outer mitochondrial membrane (OMM), each with a distinctive function and structure, which parallels their dual role as cellular powerhouses and signaling organelles. The inner mitochondrial membrane divides the mitochondrial lumen into two parts: the inner border membrane, which runs parallel to the OMM, and the cristae, which are deeply twisted, multinucleated invaginations that give room for surface area enlargement and house the mitochondrial respiration apparatus. The outer mitochondrial membrane, on the other hand, is soft and permeable. It, therefore, acts as a foundation for cell signaling pathways to congregate, be deciphered, and be transported into mitochondria. Furthermore, the OMM connects to other cellular organelles, such as the endoplasmic reticulum (ER), lysosomes, endosomes, and the plasma membrane. Mitochondria play a wide range of roles in cell biology, which is reflected in their morphological diversity. Ever since the beginning of the mitochondrial study, it has been well documented that mitochondria can have a variety of forms, with both their general and ultra-structural morphology varying greatly among cells, during the cell cycle, and in response to metabolic or cellular cues. Mitochondria can exist as independent organelles or as part of larger systems; they can also be unequally distributed in the cytosol through regulated mitochondrial transport and placement to meet the cell's localized energy requirements. Mitochondrial dynamics refers to the adaptive and variable aspect of mitochondria, including their shape and subcellular distribution.[36]
Autophagy
[edit]Autophagy is a self-degradative mechanism that regulates energy sources during growth and reaction to dietary stress. Autophagy also cleans up after itself, clearing aggregated proteins, cleaning damaged structures including mitochondria and endoplasmic reticulum and eradicating intracellular infections. Additionally, autophagy has antiviral and antibacterial roles within the cell, and it is involved at the beginning of distinctive and adaptive immune responses to viral and bacterial contamination. Some viruses include virulence proteins that prevent autophagy, while others utilize autophagy elements for intracellular development or cellular splitting.[38] Macro autophagy, micro autophagy, and chaperon-mediated autophagy are the three basic types of autophagy. When macro autophagy is triggered, an exclusion membrane incorporates a section of the cytoplasm, generating the autophagosome, a distinctive double-membraned organelle. The autophagosome then joins the lysosome to create an autolysosome, with lysosomal enzymes degrading the components. In micro autophagy, the lysosome or vacuole engulfs a piece of the cytoplasm by invaginating or protruding the lysosomal membrane to enclose the cytosol or organelles. The chaperone-mediated autophagy (CMA) protein quality assurance by digesting oxidized and altered proteins under stressful circumstances and supplying amino acids through protein denaturation.[39] Autophagy is the primary intrinsic degradative system for peptides, fats, carbohydrates, and other cellular structures. In both physiologic and stressful situations, this cellular progression is vital for upholding the correct cellular balance. Autophagy instability leads to a variety of illness symptoms, including inflammation, biochemical disturbances, aging, and neurodegenerative, due to its involvement in controlling cell integrity. The modification of the autophagy-lysosomal networks is a typical hallmark of many neurological and muscular illnesses. As a result, autophagy has been identified as a potential strategy for the prevention and treatment of various disorders. Many of these disorders are prevented or improved by consuming polyphenol in the meal. As a result, natural compounds with the ability to modify the autophagy mechanism are seen as a potential therapeutic option.[40] The creation of the double membrane (phagophore), which would be known as nucleation, is the first step in macro-autophagy. The phagophore approach indicates dysregulated polypeptides or defective organelles that come from the cell membrane, Golgi apparatus, endoplasmic reticulum, and mitochondria. With the conclusion of the autophagocyte, the phagophore's enlargement comes to an end. The auto-phagosome combines with the lysosomal vesicles to formulate an auto-lysosome that degrades the encapsulated substances, referred to as phagocytosis.[41]
Notable cell biologists
[edit]- Jean Baptiste Carnoy
- Peter Agre
- Günter Blobel
- Robert Brown
- Geoffrey M. Cooper
- Christian de Duve
- Henri Dutrochet
- Robert Hooke
- H. Robert Horvitz
- Marc Kirschner
- Anton van Leeuwenhoek
- Ira Mellman
- Marta Miączyńska[42]
- Peter D. Mitchell
- Rudolf Virchow
- Paul Nurse
- George Emil Palade
- Keith R. Porter
- Ray Rappaport
- Michael Swann
- Roger Tsien
- Edmund Beecher Wilson
- Kenneth R. Miller
- Matthias Jakob Schleiden
- Theodor Schwann
- Yoshinori Ohsumi
- Jan Evangelista Purkyně
-
Czech anatomist Jan Evangelista Purkyně is best known for his 1837 discovery of Purkinje cells.
-
Theodor Schwann, discoverer of the Schwann cell
-
Yoshinori Ohsumi, Nobel Prize winner for work on autophagy
See also
[edit]- The American Society for Cell Biology
- Cell biophysics
- Cell disruption
- Cell physiology
- Cellular adaptation
- Cellular microbiology
- Institute of Molecular and Cell Biology (disambiguation)
- Meiomitosis
- Organoid
- Outline of cell biology
Notes
[edit]- ^ Alberts, Bruce; Johnson, Alexander D.; Morgan, David; Raff, Martin; Roberts, Keith; Walter, Peter (2015). "Cells and genomes". Molecular Biology of the Cell (6th ed.). New York, NY: Garland Science. pp. 1–42. ISBN 978-0815344322.
- ^ Bisceglia, Nick. "Cell Biology". Scitable. www.nature.com.
- ^ a b c Gupta, P. (1 December 2005). Cell and Molecular Biology. Rastogi Publications. p. 11. ISBN 978-8171338177.
- ^ Hooke, Robert (September 1665). Micrographia.
- ^ a b Chubb, Gilbert Charles (1911). . In Chisholm, Hugh (ed.). Encyclopædia Britannica. Vol. 7 (11th ed.). Cambridge University Press. p. 710.
- ^ Paez-Espino D, Eloe-Fadrosh EA, Pavlopoulos GA, Thomas AD, Huntemann M, Mikhailova N, Rubin E, Ivanova NN, Kyrpides NC (August 2016). "Uncovering Earth's virome". Nature. 536 (7617): 425–30. Bibcode:2016Natur.536..425P. doi:10.1038/nature19094. PMID 27533034. S2CID 4466854.
- ^ Lavanya, P. (1 December 2005). Cell and Molecular Biology. Rastogi Publications. p. 11. ISBN 978-8171338177.
- ^ a b c d e f Cooper, Geoffrey M. (2000). "Tools of Cell Biology". The Cell: A Molecular Approach. 2nd Edition.
- ^ McKinnon, Katherine M. (21 February 2018). "Flow Cytometry: An Overview". Current Protocols in Immunology. 120 (1): 5.1.1–5.1.11. doi:10.1002/cpim.40. ISSN 1934-3671. PMC 5939936. PMID 29512141.
- ^ Doble, Mukesh; Gummadi, Sathyanarayana N. (5 August 2010). Biochemical Engineering. New Delhi: Prentice-Hall of India Pvt.Ltd. ISBN 978-8120330528.
- ^ Kaneshiro, Edna (2 May 2001). Cell Physiology Sourcebook: A Molecular Approach (3rd ed.). Academic Press. ISBN 978-0123877383.
- ^ Levetin, Estelle; McMahon, Karen (16 October 2014). Ebook: Plants and Society. McGraw Hill. p. 135. ISBN 978-0-07-717206-0.
- ^ a b c d Nelson, Daniel (22 June 2018). "The Difference Between Eukaryotic And Prokaryotic Cells". Science Trends. doi:10.31988/scitrends.20655. S2CID 91382191.
- ^ Griffiths, Anthony J.F.; Miller, Jeffrey H.; Suzuki, David T.; Lewontin, Richard C.; Gelbart, William M. (2000). "Bacterial conjugation". An Introduction to Genetic Analysis. 7th Edition.
- ^ Elosegui-Artola A, Andreu I, Beedle AE, Lezamiz A, Uroz M, Kosmalska AJ, et al. (November 2017). "Force Triggers YAP Nuclear Entry by Regulating Transport across Nuclear Pores". Cell. 171 (6): 1397–1410.e14. doi:10.1016/j.cell.2017.10.008. PMID 29107331.
- ^ "Nucleus". Genome.gov. Retrieved 27 September 2021.
- ^ "Endoplasmic Reticulum (Rough and Smooth) | British Society for Cell Biology". Retrieved 6 October 2019.
- ^ Studios, Andrew Rader. "Biology4Kids.com: Cell Structure: Endoplasmic Reticulum". www.biology4kids.com. Retrieved 27 September 2021.
- ^ "Powerhouse of the cell has self-preservation mechanism". EurekAlert!. Retrieved 27 September 2021.
- ^ Pelley, John W. (2007), "Citric Acid Cycle, Electron Transport Chain, and Oxidative Phosphorylation", Elsevier's Integrated Biochemistry, Elsevier, pp. 55–63, doi:10.1016/b978-0-323-03410-4.50013-4, ISBN 9780323034104
- ^ Cooper, Geoffrey M. (2000). "The Golgi Apparatus". The Cell: A Molecular Approach. 2nd Edition.
- ^ Verity, M A. Lysosomes: some pathologic implications. OCLC 679070471.
- ^ "Ribosome | cytology". Encyclopedia Britannica. Retrieved 27 September 2021.
- ^ Cooper, Geoffrey M (2000). The Cell: A Molecular Approach. ASM Press. ISBN 9780878931064.
- ^ Cooper, Geoffrey M. (2000). "Transport of Small Molecules". The Cell: A Molecular Approach. 2nd Edition.
- ^ "What Are the Main Functions of Cilia & Flagella?". Sciencing. Retrieved 23 November 2020.
- ^ a b Ahmad, Maria; Kahwaji, Chadi I. (2019), "Biochemistry, Electron Transport Chain", StatPearls, StatPearls Publishing, PMID 30252361, retrieved 20 October 2019
- ^ Schlessinger, Joseph (October 2000). "Cell Signaling by Receptor Tyrosine Kinases". Cell. 103 (2): 211–225. doi:10.1016/s0092-8674(00)00114-8. ISSN 0092-8674. PMID 11057895. S2CID 11465988.
- ^ Shackelford, R E; Kaufmann, W K; Paules, R S (February 1999). "Cell cycle control, checkpoint mechanisms, and genotoxic stress". Environmental Health Perspectives. 107 (suppl 1): 5–24. Bibcode:1999EnvHP.107S...5S. doi:10.1289/ehp.99107s15. ISSN 0091-6765. PMC 1566366. PMID 10229703.
- ^ a b Bernstein C, Bernstein H, Payne C. Cell immortality: maintenance of cell division potential. Prog Mol Subcell Biol. 2000;24:23-50. doi:10.1007/978-3-662-06227-2_2. PMID 10547857.
- ^ Avise JC. Perspective: The evolutionary biology of aging, sexual reproduction, and DNA repair. Evolution. 1993 Oct;47(5):1293-1301. doi:10.1111/j.1558-5646.1993.tb02155.x. PMID 28564887.
- ^ "The Cell Cycle - Phases - Mitosis - Regulation". TeachMePhysiology. Retrieved 7 October 2021.
- ^ "What is Pathology?". News-Medical.net. 13 May 2010. Retrieved 21 September 2021.
- ^ Nurse, Paul (7 January 2000). "A Long Twentieth Century of the Cell Cycle and Beyond". Cell. 100 (1): 71–78. doi:10.1016/S0092-8674(00)81684-0. ISSN 0092-8674. PMID 10647932. S2CID 16366539.
- ^ Cimprich, Karlene A.; Cortez, David (August 2008). "ATR: an essential regulator of genome integrity". Nature Reviews Molecular Cell Biology. 9 (8): 616–627. doi:10.1038/nrm2450. ISSN 1471-0080. PMC 2663384. PMID 18594563.
- ^ a b c Giacomello, Marta; Pyakurel, Aswin; Glytsou, Christina; Scorrano, Luca (18 February 2020). "The cell biology of mitochondrial membrane dynamics". Nature Reviews Molecular Cell Biology. 21 (4): 204–224. doi:10.1038/s41580-020-0210-7. ISSN 1471-0072. PMID 32071438. S2CID 211170966.
- ^ You, Zhongsheng; Bailis, Julie M. (July 2010). "DNA damage and decisions: CtIP coordinates DNA repair and cell cycle checkpoints". Trends in Cell Biology. 20 (7): 402–409. doi:10.1016/j.tcb.2010.04.002. ISSN 0962-8924. PMC 5640159. PMID 20444606.
- ^ Glick, Danielle; Barth, Sandra; Macleod, Kay F. (3 February 2010). "Autophagy: cellular and molecular mechanisms". The Journal of Pathology. 221 (1): 3–12. doi:10.1002/path.2697. ISSN 0022-3417. PMC 2990190. PMID 20225336.
- ^ Yoshii, Saori R.; Mizushima, Noboru (28 August 2017). "Monitoring and Measuring Autophagy". International Journal of Molecular Sciences. 18 (9): 1865. doi:10.3390/ijms18091865. ISSN 1422-0067. PMC 5618514. PMID 28846632.
- ^ Perrone, Lorena; Squillaro, Tiziana; Napolitano, Filomena; Terracciano, Chiara; Sampaolo, Simone; Melone, Mariarosa Anna Beatrice (13 August 2019). "The Autophagy Signaling Pathway: A Potential Multifunctional Therapeutic Target of Curcumin in Neurological and Neuromuscular Diseases". Nutrients. 11 (8): 1881. doi:10.3390/nu11081881. ISSN 2072-6643. PMC 6723827. PMID 31412596.
- ^ Levine, Beth; Kroemer, Guido (11 January 2008). "Autophagy in the Pathogenesis of Disease". Cell. 132 (1): 27–42. doi:10.1016/j.cell.2007.12.018. ISSN 0092-8674. PMC 2696814. PMID 18191218.
- ^ "Research - IIMCB". www.iimcb.gov.pl.
References
[edit]- Penner-Hahn, James E. (2013). "Chapter 2. Technologies for Detecting Metals in Single Cells. Section 4. Intrinsic X-Ray Fluorescence". In Bani, Lucia (ed.). Metallomics and the Cell. Metal Ions in Life Sciences. Vol. 12. Springer. pp. 15–40. doi:10.1007/978-94-007-5561-1_2 (inactive 31 October 2024). ISBN 978-94-007-5560-4. PMID 23595669.
{{cite book}}: CS1 maint: DOI inactive as of October 2024 (link)electronic-book ISBN 978-94-007-5561-1 ISSN 1559-0836electronic-ISSN 1868-0402 - Cell and Molecular Biology by Karp 5th Ed., ISBN 0-471-46580-1
 This article incorporates public domain material from Science Primer. NCBI. Archived from the original on 8 December 2009.
This article incorporates public domain material from Science Primer. NCBI. Archived from the original on 8 December 2009.
External links
[edit] Media related to Cell biology at Wikimedia Commons
Media related to Cell biology at Wikimedia Commons- Aging Cell
- "Francis Harry Compton Crick (1916–2004)" by A. Andrei at the Embryo Project Encyclopedia
- "Biology Resource By Professor Lin."




